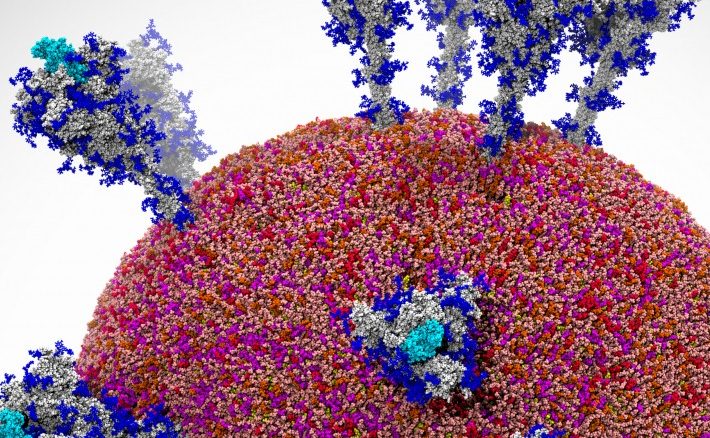
For more than three decades, researchers have used a particular simulation method for molecular dynamics called Ab initio molecular dynamics, or AIMD, which has proven itself to be the method most accurate for analyzing how atoms and molecules move and interact over a fixed time period. In this time of a global COVID-19 pandemic that continues to rage in the United States and elsewhere, molecular dynamics is a key technology in the research and development of therapeutics and vaccines to combat the deadly coronavirus.
However, while the precision of the AIMD approach has made it the preferred method over Standard Molecular Dynamics (SMD), there have been limitations. In particular, the level of precision that makes AIMD so effective also requires huge amounts of computation capabilities, which has limited the size of the systems it can used to study. While molecular dynamics is used to get a clear sense of how a system – such as a single cell or a cloud of gas – progresses over time, AIMD could only be used to small systems with a maximum size of thousands of atoms.
That said, a team of eight scientists from both the United States and China, using the “Summit” supercomputer at the Oak Ridge National Laboratory, has developed a new protocol based on machine learning techniques that it says can simulate the movements of more than 100 million atoms per day over more than a nanosecond-long trajectory with ab initio accuracy.
The development of the Deep Potential Molecular Dynamics (DPMD) protocol was awarded the ACM Gordon Bell Prize winner at the virtual SC20 supercomputer event last week, besting five other competitors for the award. The show was a digital event this year because of the coronavirus outbreak, so the ACM Gordon Bell organization also gave a prize dedicated to research into COVID-19, the disease caused by the coronavirus.
In a detailed paper, the DPMD team wrote about the limitations of the AIMD method, saying that the “computational cost of AIMD generally scales cubically with respect to the number of electronic degrees of freedom. On a desktop workstation, the typical spatial and temporal scales achievable by AIMD are ∼100 atoms and ∼10 picoseconds. From 2006 to 2019, the peak performance of the world’s fastest supercomputer has increased about 550-folds, (from 360 TFLOPS of BlueGene/L to 200 PFLOPS of Summit), but the accessible system size has only increased 8 times (from 1K Molybdenum atoms with 12K valence electrons to 11K Magnesium atoms with 105K valence electrons), which obeys almost perfectly the cubic-scaling law. Linear-scaling DFT methods have been under active developments, yet the pre-factor in the complexity is still large, and the time scales attainable in MD simulations remain rather short.”
The problem is that for such simulations as complex chemical reactions, electrochemical cells, nanocrystalline materials and radiation damage, what’s needed are systems sizes that range from thousands to hundreds of millions of atoms and time scales up to a microsecond or more, which is far beyond what can be done with AIMD, they wrote.
Molecular dynamics schemes based on machine learning models trained with ab initio data in recent years held the promise of boosting AIMD and the Deep Potential protocol has shown the ability to offer accuracy comparable to AIMD and efficiency close to EFF (empirical force fields)-based molecular dynamics.
“The accuracy of the DP [Deep Potential] model stems from the distinctive ability of deep neural networks (DNN) to approximate high-dimensional functions, the proper treatment of physical requirements like symmetry constraints, and the concurrent learning scheme that generates a compact training dataset with a guarantee of uniform accuracy within the relevant configuration space,” the researchers wrote.
The IBM-built Summit system is the world’s second-fastest supercomputer, according to the biannual Top500 list, the latest of which was released at SC20. It delivers a performance of 148.8 petaflops and comprises 4,356 nodes, each one holding two 22-core IBM Power9 processors and six Tesla V100 GPUs from Nvidia. It had topped the list from November 2018 to June, when it was displaced by the Fugaku system in Japan, powered by Fujitsu’s Arm-based A64FX processors. The system runs on more than 7.6 million cores.
The DPMD researchers said they wanted to leverage the GPUs on Summit to run almost all of the computational tasks and many of the communications tasks. However, because of relatively limited size of the computational granularity in the Deep Potential model, relying heavily on the GPUs would be inefficient. To get past that hurdle, the team made a range of algorithmic innovations, including introducing a new data layout for the neighbor list that avoided branching in the computation of the embedded matrix – and thus increasing the computational granularity of DeepMD implementation. (The DeepMD scheme was implemented in an open-source package called DeePMD-kit.)
Other efforts included compressing the elements in the new data structure into 64-bit integers to improve GPU optimization for the customized TensorFlow operations and creating mixed-precision computation for the Deep Potential model.
The researchers said their work “provides a vivid demonstration of what can be achieved by integrating physics-based modeling and simulation, machine learning, and efficient implementation on the next-generation computational platform. It opens up a host of new exciting possibilities in applications to material science, chemistry, and biology. It also poses new challenges to the next-generation supercomputer for a better integration of machine learning and physical modeling. We believe that this work may represent a turning point in the history of high-performance computing, and it will have profound implications not only in the field of molecular simulation, but also in other areas of scientific computing.”
It also will have an impact as the world enters the exascale computing era, they wrote. There’s been rapid innovation in many-core architectures, memory bandwidth and other areas. However, the researchers wrote, such innovation “essentially requires a revisit of the scientific applications and a rethinking of the optimal data layout and MPI communication at an algorithmic level, rather than simply offloading computational intensive tasks. In this paper, the critical data layout in DeePMD is redesigned to increase the task granularity, then the entire DeePMD-kit code is parallelized and optimized to improve its scalability and efficiency on the GPU supercomputer Summit. The optimization strategy presented in this paper can also be applied to other many-core architectures.”
That includes converting it for the “Frontier” supercomputer, which will run the Heterogeneous-compute Interface for Portability (HIP) programming model, which will be based on GPUs from AMD. Frontier, which will be based on compute technologies from Cray (now owned by Hewlett Packard Enterprise following its $1.3 billion acquisition last year) and AMD.
The ACM said in a statement that those winning the Gordon Bell Prize need to show that their proposed algorithm can scale and run efficiently on the world’s most powerful supercomputers. This year’s winners “developed a highly optimized code (GPU Deep MD-Kit), which they successfully ran on the Summit supercomputer. The team’s GPU DeepMD-Kit efficiently scaled up to the entire Summit supercomputer, attaining 91 PFLOPS … in double precision (45.5 percent of the peak) and 162/275 PFLOPS in mixed-single/half precision,” it said.




Be the first to comment