Since late February and early March, when the coronavirus outbreak began to spill out of China in earnest and spread its sickness and death across Europe, throughout the United States, and into other parts of the globe, IT companies have stepped up to lend their technologies to fight the pandemic.
Vendors have offered access to their products and services on their own or join together, pooling their resources and capabilities. For some it’s a push aimed at Covid-19 in particular; for others, an extension of programs already underway designed to use technology to address global concerns, such as climate change.
Late last month we wrote about the formation of the Covid-19 High Performance Computing Consortium, an organization comprising IBM, Microsoft, Google and Amazon – and more recently, Nvidia – as well as government organizations like the National Science Foundation and Department of Energy (DoE) pooling the aggregate computing power of 16 supercomputers at the national laboratories (including Summit, the massive IBM-based supercomputer at the Oak Ridge National Lab) and facilities like the Texas Advanced Computing Center (TACC). The consortium is giving medical researchers and scientists access to a combined 330 petaflops of compute power to get a better understanding of the coronavirus as well as possible treatments and vaccines.
Soon after Microsoft, Facebook, Twitter and other tech vendors announced they were partnering with the World Health Organization (WHO) on the #BuildforCOVID19 Global Online Hackathon, a software competition aimed at accelerating the development of software solutions targeting challenges related to the pandemic, from health initiatives to economics and education. The idea was to pick the best proposals and then have the developer work with engineers from Facebook, Microsoft, Slack, Twitter and others to develop the software. Similar hackathons have been launched around the world, including by MIT – dubbed MIT COVID-19 Challenge: Beat the Pandemic – over the weekend of April 4 and 5.
There are myriad other examples, such as Core Scientific, an expert in infrastructure and software for artificial intelligence (AI) and blockchain, giving coronavirus researchers access to a cloud-based infrastructure that includes NetApp’s OnTap AI solution and flash-based storage and Core Scientific’s Plexus AI stack, and which is powered by Nvidia’s DGX systems, and Aruba Networks, owned by Hewlett Packard Enterprise, offering wireless networking bundles and volunteers to hospitals. Rivals Apple and Google announced April 10 that they are working together on a solution that will include APIs and OS-level technology to enable contact tracing, a key step in establishing the degree that the coronavirus has spread in a community or country.
In late February, IBM turned the focus of its Call for Code challenge to Covid-19. Call for Code, now in its third year, was launched by IBM and The David Clark Foundation to encourage developers to use their skills to create software to address global challenges, such as natural disasters and climate change. Now the challenge is the pandemic.
“In a very short period of time, COVID-19 has revealed the limits of the systems we take for granted,” Willie Tejada, chief developer advocate at IBM, wrote in a blog post March 20, explaining the need for an accelerated timeline. The initial deadline for submissions is April 27, with the top three solutions being announced May 5, with IBM working with those developers to get their offerings into the field. The company also will keep the track open through July to get more solutions in.
Graphs4Good
Similarly, graph database vendor Neo4j for several years has run its Graphs4Good program aimed at encouraging users to develop and connect graph-powered projects that address global challenges, from promoting positive social change to upholding democratic values. Graph databases have been around for years – Neo4j launched its first iteration in 2007 – as a way of querying massive data lakes and data warehouses in the growing big data and analytics scene. There are a growing number of graph database vendors and open source projects, from TigerGraph, GraphDB, and Titan. Proponents see graph databases as a more logical way than relational and other database technologies to query huge pools of data, focusing on the idea of connections between data points. It uses nodes – data pieces like people, places or categories – and edges to find relationships within the data.
Probably the highest profile Graphs4Good project was run by the International Consortium of Investigative Journalists (ICIJ), which has been using Neo4j technology since 2015. In 2016, the organization used the technology with the Pulitzer Prize-winning Panama Papers effort, which included 11.5 million leaked documents total 2.6 TB of data dating back to the 1970s that uncovered massive illegal offshore business fraud. Beyond journalism, the Graphs4Good project has focused on healthcare (cancer and diabetes), space exploration (NASA), the humanities (medieval manuscripts and historical Renaissance artifacts) and women in tech.
“We like supporting projects that make use of connected data for the greater good,” Alicia Frame, lead project manager and data scientist at Neo4j, tells The Next Platform. “So you see things like the Panama Papers database. It’s really important to us as a company to use our technology not just to help the big banks fight fraud or make better recommendations, but also to do good in the world.”
Turning The Focus to COVID-19
Over the last two months, developers began telling Neo4j about coronavirus-related projects they had in mind but couldn’t afford to get them off the ground. Given the interest, the company turned the focus of its Graphs4Good program to COVID-19, sponsoring a hackathon that’s running through April 14 for such projects that looks at everything rom contact tracing and drug discovery to creating a central place for information sharing, Frame says.

“If someone is doing a project, we want to support them,” she says. “Is that giving them an enterprise license for free? Is it making credits available to running on a cloud platform? Is it giving them early access to that graph data science library or is it giving them internal resources? How do you capture all this energy and let people work together, but also through empowering projects that actually have gained traction and they’re trying to scale up and provide something publicly?”
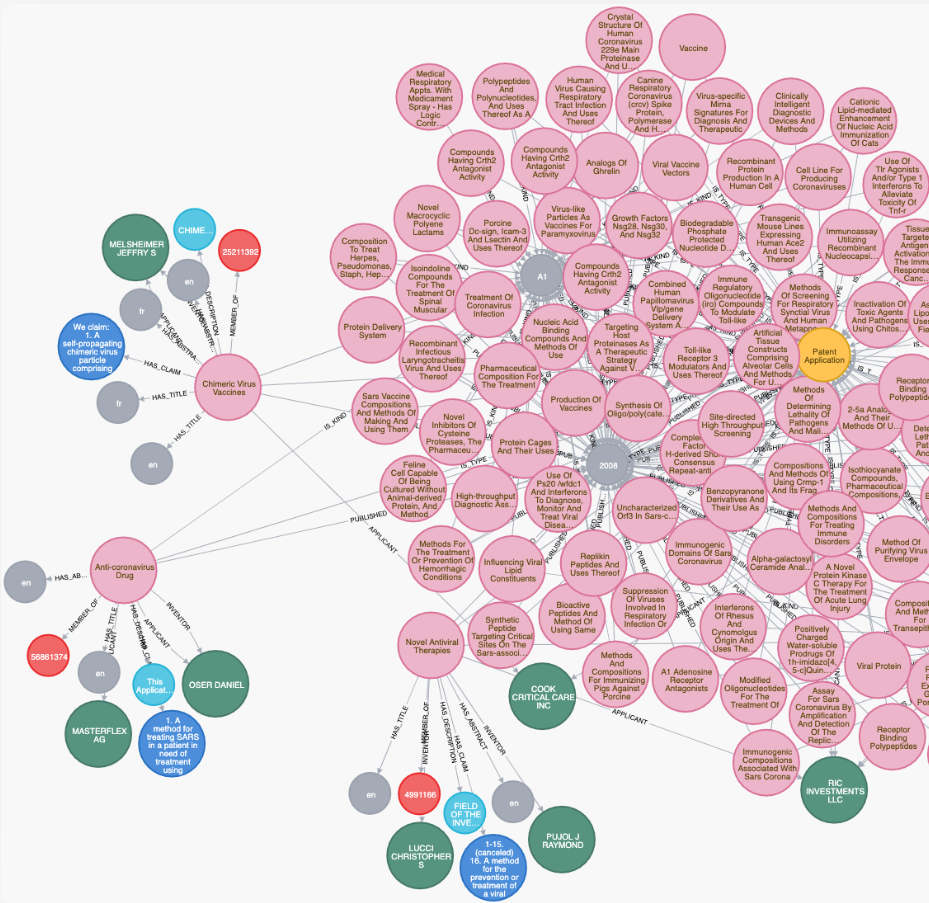
One project underway using the Neo4j technology is being headed by the German Center for Diabetes Research (DZD). The organization is one of six nonprofits in the country that study widespread diseases like diabetes, cancer and infectious diseases, trying to determine not only what causes these diseases but also any connections between them, Alexander Jarasch, head of data knowledge and management at the DZD, tells The Next Platform. The organizations use Neo4j’s graph technology to help them sort through and query tens of thousands of pages of literature on these diseases stored in a massive database. Jarasch got the idea to use natural language processing to automatically understand and make sense of the text data and to help build a knowledge graph out of 32 million articles, and then be able to query the data about connections between such areas as genes, proteins, symptoms and side effects, as well as between diseases.
Now Jarasch and other researchers, volunteers and companies – including Neo4j – are using these capabilities in building a knowledge graph for Covid-19, an effort that began almost a month ago. The goal is to build a resource that integrates public datasets about the coronavirus that researchers and scientists more quickly find the needed information from such sources publications, case statistics, genes and molecular data. Jarasch estimates that over the past three months, there are have been about 45,000 publications about Covid-19 – primarily out of Asia, where the outbreak began – some of which haven’t been peer reviewed, others that are essentially summaries and some that are little more than bits and pieces of information.

“This is where we use Ne04j as a graph database underneath, to have the bits and pieces collected,” he says. Then they not only want to have “the publications connected, but with the relevant biological entities, like genes and proteins, as we do it for diabetes. Then we want to do an automated text analysis and find things – say genes – that are mentioned in one of the other publications. This is the first step of our project and where we provide researchers and interested people a database where we not only have information from publications, but also from biological entities. We had another idea of companies who could provide us with relevant information and this is when you think about patents and intellectual property. When we think about ten years back, there was the SARS virus, which was also very critical [and] was also a coronavirus and there were a bunch of patents that have been filed for devices for ventilators or for drugs [or] vaccinations and I think that in these patent text, there’s a big potential that we can analyze in order to find maybe a drug that is similar and that might work or might be repurposed for COVID-19. This is the reason why we also integrated about 16,000 patents that are somehow relevant to coronaviruses in general. This summed up to a huge graph database with Neo4j, where we have the database and several companies joined us in these initiatives and also from the graph family.”
The project uses Python scripting to help integrate data and a range of other companies, such as yWorks and Graphelion for visualization, Structr and its graph-based application platform, and GitHub as a code repository, Jarasch says. It also uses data from some relational databases. Over the next two weeks, one part of the team will apply machine learning techniques to find patterns in the data. At the same time, other parts of the team comprising infectious disease researchers will help to determine what information they should search for in the huge amounts of data, such as genes, symptoms or results from clinical trials, he says.
“In theory, everything you do with graph databases, you could potentially do in SQL or in relational databases,” Jarasch says. “But it would be much more complicated and the runtime of the same query would take a factor of 1,000 or even a million [more], so we are using graph databases because our field of interest is highly connected. Everything is connected. A gene is connected to a protein, a protein to a phenotype, a phenotype to a disease. The disease is connected to another disease. The kind of data we have is highly connected, so it makes much sense to use a native graph database.”
He also notes that graphs are common in such research. When someone is asked to draw a mode of action regarding a drug or a molecular pathway in biology, they usually do so in graphs, he says, adding that “in terms of runtime graph databases, they can grow size-wise and data-wise. They can grow in size as the data grows. But the query in terms of what the data you touch now is only a small portion of it. That would be not feasible with the relational database.”
Neo4j’s Frame says graph databases lend themselves to such research.
“When we talk about graph databases, the idea is instead of rows and columns … like in SQL land, you have nodes and relationships,” she says. “You have the connection between your data points on equal footing with the data points themselves. That sounds kind of like a niche use case until you start to think about, ‘How do I represent things like contact tracing?’ It’s ‘Oh, I have people interacting with each other.’ Or doing something around supply chain, where you are asking what are the resources, what are the bottlenecks, where are the redundancies? All of a sudden, you’re like, ‘Oh, wait, this is a graph model.’ Here’s the manufacturer. It goes from here to here. There are these multiple relationships to get these widgets in this factory. The third use case we’ve seen a lot of is this knowledge graph pharma R&D. All of these are really naturally quite well represented as a graph.”
It’s easier to pull insights about the global topology, essentially clustering ideas together, Frame says. Instead of searching based on similar descriptors, “I can say, ‘This paper has these two genes that regulate each other and the third paper that I found has the gene that’s interacting with the regulated gene.’ Then I can start to connect the concepts very naturally. I can start running algorithms across all of these connected concepts and understand the topology without combing through everything one thing at a time.”



Be the first to comment